| International Journal of Clinical Pediatrics, ISSN 1927-1255 print, 1927-1263 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, Int J Clin Pediatr and Elmer Press Inc |
| Journal website http://www.theijcp.org |
Original Article
Volume 3, Number 1, March 2014, pages 12-15
Coding Regions of MSX1 do not Contribute to Non-Syndromic Cleft Lip With/Without Palate in Turkish Patients
Korkut Ulucana, i, Arzu Akcayb, Burak Aksoyc, Mehmet Boyrazd, Deniz Kirace, Deniz Ergecf, Necati Taskıng, Ozhan Ozcelebilerc, Muhsin Konuka, Teoman Akcayh, A. Ilter Guneyf
aDepartment of Molecular Biology and Genetics, Faculty of Engineering and Natural Sciences, Uskudar University; Turkey
bUnit of Bone Marrow Transplantation, Bahcelievler Madical Park Hospital, Istanbul, Turkey
cDepartment of Reconstructive Surgery, Faculty of Medicine, Marmara University, Istanbul, Turkey
dDivision of Pediatric Endocrinology, Faculty of Medicine, Fatih University, Ankara, Turkey
eDepartment of Medical Biology, Faculty of Medicine, Yeditepe University, Istanbul, Turkey
fDepartment of Medical Genetics, Faculty of Medicine, Marmara University, Istanbul, Turkey
gDivision of Pediatrics, Kanuni Sultan Suleyman Education and Research Hospital, Turkey
hDivision of Pediatric Endocrinology, Kanuni Sultan Suleyman Education and Research Hospital, Istanbul, Turkey
iCorresponding author: Korkut Ulucan, Uskudar University, Faculty of Engineering and Natural Sciences, Department of Molecuar Biology and Genetics, Haluk Turksoy Sok. No:14, Altunizade, 34662, Uskudar, Istanbul, Turkey
Manuscript accepted for publication January 20, 2014
Short title: MSX1 Determination in Turkish Patients
doi: https://doi.org/10.14740/ijcp136e
| Abstract | ▴Top |
Background: One of the candidate genes for non-syndromic cleft lip with/without palate (NS-CL/P) is muscle segment homeobox 1 (MSX1) gene. MSX1 codes for a homeodomain protein that function as a transcriptional repressor during craniofacial development. In this study, we investigated the contribution of coding regions and exonic-intronic boundaries of MSX1 in Turkish NS-CL/P patients and the gender distribution of NS-CL/P in examined cohort.
Methods: We analyzed a total of 100 NS-CL/P Turkish patients, of whom 61 were boy and, 100 non-affected children of whom 44 were boys. Genotyping procedure was assessed by polymerase chain reaction-sequencing protocol. Fisher Exact Analysis was performed to compare the gender difference.
Results: We detected no variations in coding regions and intronic-exonic boundaries of MSX1 in both groups. Boys to girls’ ratio were 1.6 in our patient group. There was a statistically significant gender difference between groups, boys were more effected than girls (P = 0.023).
Conclusions: This was the first report including MSX1 in Turkish patients with NS-CL/P. We detected no sequence variations in the coding regions, including the exonic-intronic boundaries of MSX1 in our study population. More comprehensive analysis of MSX1 in Turkish population is required to fulfill MSX1 role for the onset of the anomaly.
Keywords: Cleft lip with/without palate; Congenital; MSX1; Multifactorial; Orofacial clefts
| Introduction | ▴Top |
Non-syndromic cleft lip with/without palate (NS-CL/P) (MIM 119530) affects about 1/1,000 live births, and shows wide variability related to geographic distribution, ethnic background and socioeconomic status [1]. Gene-gene and gene-environmental interactions have been implicated in NS-CL/P and several different loci and genes have been associated with NS-CL/P. Studies including monozygotic and dizygotic twins revealed the genetic factors as strong components of orofacial clefts, concordance rate was reported as 40-60% and 3-5%, for monozygotic and dizygotic twins, respectively [2]. Lack of 100% concordance in monozygotic twins suggests the environmental factors importance, not the individual’s genotype alone is responsible for the formation of orofacial clefts [3].
Recent studies tried to identify the candidate gene functions in orofacial clefts in different populations and the candidate gene list includes the genes like IRF6, muscle segment homeobox 1 (MSX1), TGFB3, FOXE1, FGFR1, FGFR2, FGF8, PDGFC, CRISPLD2, PVRL1, GABRB3, MSX2, SATB2, TBX10, TBX22, GLI2, JAG2, MTHFR, RARA, LHX8, SKI and SPRY2 [3]. The great majority and the common conclusions of the studies are no single locus has been clearly emerged as necessary for the onset of NS-CL/P.
MSX1 gene is a member of MSX family and consists of two exons and one intron. Gene encodes a homeodomain protein that functions as a transcriptional repressor during embryogenesis and systems development like craniofacial, limb and nervous [4]. Studies in mice and humans supported the MSX1 as a strong candidate gene in orofacial clefts. Mice that are homozygous for MSX1-null alleles die right after birth and have severe craniofacial abnormalities including cleft palate [5]. Also, mice and humans lacking MSX1 function show non-syndromic clefts of the secondary palate and tooth agenesis [6].
In this study, we aimed to determine the sequence variations in the coding regions of MSX1 regarding to NS-CL/P and indicate the association of it and NS-CL/P that may be etiological in Turkish patients and also to identify the gender difference between patients and controls in the examined individuals.
| Materials and Methods | ▴Top |
Subjects
A total of 100 patients (aged between 4 and 7) without any family history of NS-CL/P and same number of randomly chosen age-matched controls were recruited for the study. All of the individuals enrolled in the study were of Turkish ancestry, without any immigrants. All the patients were operated at Marmara University, Faculty of Medicine, in the department of reconstructive surgery. Of the 100 patients, 61 were boys (61%) and 39 were girls (39%) whereas 44 were boys (44%) and 56 were girls (56%) in control group. Detailed information about the subjects and parents were obtained either directly or by telephone calls. Patients who had any other genetic or dental anomalies were excluded from the study. Ethical Committee of Marmara University approved the experimental protocol and the study procedure was in accordance with the principles of the Declaration of Helsinki II. Written informed consents indicating the aim and protocol of the study were taken from one of their parents who were in charge of the child.
Amplification of MSX1 gene exons
DNA from subjects were isolated from 200 µL peripheral blood by using High Pure PCR Template Preparation Kit (Roche, Germany) using the protocol outlined in the manufacturer’s guide. For the mutation detection, two exons including exon-intron boundaries of the gene were amplified. Exon 2 was amplified in two overlapping fragments (exon2.1 and 2.2) so as to take better results in sequencing analysis. Table 1 lists the primers used for amplifying procedures, Tm values and the length of the amplicons. PCR amplifications were performed in a total volume of 50 µL containing 50-100 ng DNA template in 10 mM Tris-HCl (pH 8.0), 50 mM KCl, 1.5 mM MgCl2, 100 mM each of dNTPs, 1.0 U Taq DNA polymerase and 1.0 mM of each primer. The conditions of PCR amplification were as follows: a denaturation step at 94 °C for 3 min followed by 34 cycles at 94 °C for 45 sec, Tm values listed in Table 1 for 1 min, 72 °C for 1 min, a final extension at 72 °C for 7 min. All the amplicons were visualized in 2% agarose gel with ethidium bromide (0.2 g/mL) under ultraviolet light.
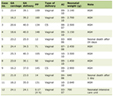 Click to view | Table 1. Primer Sequences, Tm Values and the Length of the Amplicons of the Ampolified Exons |
Sequencing of MSX1 gene
Amplicons formed by PCR reactions were sequenced by ABI PRISM 310 Genetic Analyzer (Applied Biosystems, USA) in both directions by using the same primers. Sequencing results were compared to wild type MSX1 as stated in Genbank.
Statistical analysis
Statistical analysis was assessed by using SPSS for Windows version 20.0 (SPSS inc., Chicago, IL, USA). The significance in the statistical analyses between the patient and control groups was assessed using the Fisher Exact Analysis. P < 0.05 was considered to represent a statistically significant result.
| Results | ▴Top |
PCR and sequencing results revealed no mutations in the examined regions of the genes when compared to the MSX1 wild type gene of Genbank.
Fisher Exact Analysis was performed in order to compare the gender difference and NS-CL/P. There was a statistically significant difference in genders between patients and control groups. Boys were more affected than girls (P = 0.023; P < 0.05 is accepted as sense) (Table 2). In our patient group, boys to girls’ ratio were 1.6.
 Click to view | Table 2. Sex Distribution and Statistically Comparison of Patients and Controls |
| Discussion | ▴Top |
NS-CL/P is congenital and complex anomaly that do not follow simple dominant-recessive inheritance pattern. Up to 20 genes were assumed to interact multiplicatively to cause oral clefts. Because of the genetic and ethnic heterogeneity property of the NS-CL/P, different population based studies need to be carried out including the candidate genes that are thought to be responsible for the onset of orofacial clefting.
Our study was based on case-control design rather than case-parent study design. The reason that executed us to choose this study design was not having any information about the variations of MSX1 in Turkish population. This study design was reported to be more useful in understanding the role of candidate gene in populations whose population stratification is not clear [7]. And secondly, case-control studies are more sensitive to detect the minimal changes in allele frequency between cases and controls especially when investigating complex genetic disorders and when working with small sample size.
In our study, we detected no sequence variations in the examined regions of MSX1, two exons including intronic-exonic boundaries in Turkish NS-CL/P patients. There are some reports that researchers found no MSX1 mutations in patients with clefts [8, 9]. According to the previous studies, mutations in MSX1 can contribute to NSCL/P as many as 2% of the total cases [10, 11]. Some studies reported the mutations and/or polymorphisms do not contribute to the anomaly in the examined populations [12, 13], and some others reported the variations contribute to the anomaly [14, 15]. The unrepeated results from different populations with different ethnicity showed the variability nature of MSX1 for the onset of NS-CL/P.
Due to some reasons, we were unable to detect any variations in our patients. We recruited 100 NS-CL/P unrelated patients having no family history of any genetical disease. As this anomaly is believed to be multigenic, MSX1 may not be the candidate gene for this phenotype in our patients. MSX1 protein interacts with its downstream molecules like Dlx, Lhx2, Bmps and TATA binding proteins during palatogenesis [16]. Variations in these genes and proteins can also cause orofacial clefts or variations in promoter or in intron that can affect splicing can cause anomaly, although the coding sequences are proper. Epigenetic factors, which cannot be detected by simple exon sequencing, can also alter gene function and these may be the reason why we couldn’t find an association between MSX1 and NS-CL/P in our study group.
When we compare the gender differences between patients and controls, we found that NS-CL/P was statistically higher in boys (P = 0.023). The ratio of boys to girls was 1.6 in our study cohort. Our results supported some studies that were carried out in different populations [17, 18]. They all reported male to female ratios were bigger than 1, indicating CL/P has more incidences in boys. However, Elliot et al [19] could not find any gender difference for orofacial clefts in Zambia population and Suleiman et al [20] reported a higher incidence of girls in Sudan population. It is quite difficult to make a precise decision about the gender distribution in our study, although there is a statistically difference between groups. In order to make a precise decision, extended numbers of patients and controls should be examined. One explanation for this condition may be explained as the number of the boys in patients group was higher than the girls and the number of the boys in control group was lower than the girls, although the patients and the controls were enrolled in the study randomly.
This first study, including Turkish patients and MSX1 in the etiology of NS-CL/P does not allow us to make decisive proposal. Recently we published a family having members with NS-CL/P and in this family, intronic CA repeats may be the causing factor for the onset of the anomaly [21].
It is hard to identify the candidate gene functions in multi-factorial inheritance patterns. As a result of this status, a large and different population analysis including case- control subjects are needed to identify the candidate genes’ roles for the onset of diseases. Although we detected no variations in the coding regions of MSX1, MSX1 and its interacting genes might play an important role in the etiology of orofacial clefts. Analyzing the whole gene and identifying intronic CA repeats will be the focus of our further studies.
| References | ▴Top |
- Carinci F, Scapoli L, Palmieri A, Zollino I, Pezzetti F. Human genetic factors in nonsyndromic cleft lip and palate: an update. Int J Pediatr Otorhinolaryngol. 2007;71(10):1509-1519.
doi pubmed - Jugessur A, Farlie PG, Kilpatrick N. The genetics of isolated orofacial clefts: from genotypes to subphenotypes. Oral Dis. 2009;15(7):437-453.
doi pubmed - Murray JC. Gene/environment causes of cleft lip and/or palate. Clin Genet. 2002;61(4):248-256.
doi pubmed - Catron KM, Wang H, Hu G, Shen MM, Abate-Shen C. Comparison of MSX-1 and MSX-2 suggests a molecular basis for functional redundancy. Mech Dev. 1996;55(2):185-199.
doi - Satokata I, Maas R. Msx1 deficient mice exhibit cleft palate and abnormalities of craniofacial and tooth development. Nat Genet. 1994;6(4):348-356.
doi pubmed - Vastardis H, Karimbux N, Guthua SW, Seidman JG, Seidman CE. A human MSX1 homeodomain missense mutation causes selective tooth agenesis. Nat Genet. 1996;13(4):417-421.
doi pubmed - Khoury MJ. Human genome epidemiology: translating advances in human genetics into population-based data for medicine and public health. Genet Med. 1999;1(3):71-73.
doi pubmed - Modesto A, Moreno LM, Krahn K, King S, Lidral AC. MSX1 and Orofacial Clefting with and without Tooth Agenesis. J Dental Res. 2006; 85: 542-547.
doi - De Muynck S, Schollen E, Matthijs G, Verdonck A, Devriendt K, Carels C. A novel MSX1 mutation in hypodontia. Am J Med Genet A. 2004;128A(4):401-403.
doi pubmed - Tongkobpetch S, Siriwan P, Shotelersuk V. MSX1 mutations contribute to nonsyndromic cleft lip in a Thai population. J Hum Genet. 2006;51(8):671-676.
doi pubmed - Suzuki Y, Jezewski PA, Machida J, Watanabe Y, Shi M, Cooper ME, Viet le T, et al. In a Vietnamese population, MSX1 variants contribute to cleft lip and palate. Genet Med. 2004;6(3):117-125.
doi pubmed - Vieira AR, Avila JR, Daack-Hirsch S, Dragan E, Felix TM, Rahimov F, Harrington J, et al. Medical sequencing of candidate genes for nonsyndromic cleft lip and palate. PLoS Genet. 2005;1(6):e64.
doi pubmed - Lace B, Vasiljeva I, Dundure I, Barkane B, Akota I, Krumina A. Mutation analysis of the MSX1 gene exons and intron in patients with nonsyndromic cleft lip and palate. Stomatologija. 2006;8(1):21-24.
pubmed - Jagomagi T, Nikopensius T, Krjutskov K, Tammekivi V, Viltrop T, Saag M, Metspalu A. MTHFR and MSX1 contribute to the risk of nonsyndromic cleft lip/palate. Eur J Oral Sci. 2010;118(3):213-220.
doi pubmed - Salahshourifar I, Halim AS, Wan Sulaiman WA, Zilfalil BA. Contribution of MSX1 variants to the risk of non-syndromic cleft lip and palate in a Malay population. J Hum Genet. 2011;56(11):755-758.
doi pubmed - Bendall AJ, Abate-Shen C. Roles for Msx and Dlx homeoproteins in vertebrate development. Gene. 2000;247(1-2):17-31.
doi - Li Z, Ren A, Liu J, Zhang L, Ye R, Li S. High prevalence of orofacial clefts in Shanxi Province in northern China, 2003-2004. Am J Med Genet A. 2008;146A(20):2637-2643.
doi pubmed - Rajabian MH, Aghaei S. Cleft lip and palate in southwestern Iran: an epidemiologic study of live births. Ann Saudi Med. 2005;25(5):385-388.
pubmed - Elliott RF, Jovic G, Beveridge M. Seasonal variation and regional distribution of cleft lip and palate in Zambia. Cleft Palate Craniofac J. 2008;45(5):533-538.
doi pubmed - Suleiman AM, Hamzah ST, Abusalab MA, Samaan KT. Prevalence of cleft lip and palate in a hospital-based population in the Sudan. Int J Paediatr Dent. 2005;15(3):185-189.
doi pubmed - Ulucan K, Akcay A, Taskın N, Akcay T, Konuk M. MSX1 intronic CA repeat polymorphism is associated with non-syndromic cleft lip with/without palate in a Turkish family. Dis Mol Med. 2013; 1:68-71.
doi
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
International Journal of Clinical Pediatrics is published by Elmer Press Inc.
