| International Journal of Clinical Pediatrics, ISSN 1927-1255 print, 1927-1263 online, Open Access |
| Article copyright, the authors; Journal compilation copyright, Int J Clin Pediatr and Elmer Press Inc |
| Journal website http://www.theijcp.org |
Case Report
Volume 3, Number 1, March 2014, pages 22-28
Next Generation Sequencing in Differential Diagnosis of Childhood Melanoma From Spitz Nevus: A Case Report
Xun Wua, b, Keith Tomaszewicza, Lloyd Hutchinsona, Ediz Cosara, April Denga
aDepartment of Pathology, University of Massachusetts Medical School, UMass Memorial Medical Center, Worcester, MA 01605, USA
bCorresponding author: Xun Wu, Department of Pathology, University of Massachusetts Medical School, Biotech 3, One Innovation Drive, Worcester, MA 01605, USA
Manuscript accepted for publication March 20, 2014
Short title: Childhood Melanoma from Spitz Nevus
doi: https://doi.org/10.14740/ijcp148e
| Abstract | ▴Top |
Melanomas are uncommon in children and share many of the characteristics of benign Spitz nevi in their clinical presentation as well as histopathology. Therefore, the diagnosis could be challenging. Currently, molecular diagnostic tools are being explored to aid the diagnosis in the pediatric melanoma. Melanomas often harbor multiple gene mutations and gene number variations, including BRAF (7q34), CCND1 (11q13), RREB1 (6p25), Cep 6, MYB (6q23), PIK3CA (3q26), PTEN (10q23), CDKN2A (9p21), c-KIT (4q12) and MYC (8q24), but these changes are rare in benign melanocytic nevi, including Spitz nevus of which the signature gene abnormalities had been recently revealed. Next generation sequencing (NGS) is a new molecular technique allowing the detection of a broad spectrum of genome abnormalities simultaneously, thus providing a potential powerful tool for assisting the diagnosis of childhood melanoma. We herein present a 12-year-old female with a melanocytic neoplasm showing histopathologic features highly suggestive of malignant melanoma. A variety of molecular analysis studies were employed including NGS, which revealed BRAF mutation, BRAF copy number addition, and CDKN2A and PTEN deletion. Fluorescence in situ hybridization studies were also used, which not only confirmed the gene number variation in BRAF, CDKN2A and PTEN, but also revealed gain in MYC. These results added valuable information in the diagnosis of melanoma. This case demonstrated the usefulness of molecular analysis as the ancillary tools in assisting the diagnosis of childhood melanoma.
Keywords: Malignant melanoma; Spitz nevus; Gene mutation; Next generation sequencing; Fluorescence in situ hybridization
| Introduction | ▴Top |
Malignant melanoma is the most deadly skin cancer. There are more than 70,000 new cases resulting in more than 9,000 deaths each year in the United States [1]. However, melanoma in pediatric population is rare, accounting for less than 1% of all melanoma cases with only 300-400 new diagnoses each year, rarely metastasizing or resulting in death [2-5]. On the other hand, Spitz nevi, a variant of benign melanocytic nevi, are commonly seen in children. Spitz nevi often show clinical and histopathologic features that are shared by both benign nevi and malignant melanomas, thus posting a notorious diagnostic challenge [4, 6, 7].
For melanoma patients, early detection and treatment is the key to achieve good prognosis as the survival rate is slim for advanced melanoma. Failing to recognize the malignancy can lead to inadequate therapy and death, while a false positive diagnosis can cause inappropriate over-treatment such as lymph node dissection and chemotherapy as well as psychological stress on the patient and family, setting aside the financial burden to the health care system. Pediatric melanoma can often be misdiagnosed as benign nevus due to its rarity and resemblance to benign Spitz nevus, and the fact that physicians are reluctant to perform biopsy in children [8]. Even if the biopsies are sometimes performed, owing to the overlap histopathologic features between melanoma and benign nevus, misdiagnosis is often. One study showed that up to 40% of children originally diagnosed with Spitz nevi were later found to have melanomas [5]. Some other studies even established metastasis as the only gold standard for the diagnosis of malignancy [5].
Currently the utility of molecular tools for the diagnosis of pediatric melanoma is being explored. Detection of an oncogenic mutation by itself is usually insufficient to make a diagnosis of malignant melanoma since nevi often harbor the same mutations (for example BRAF) [9]. Increasing evidence shows that melanomas often harbor multiple gene abnormalities while benign nevi have none or fewer ones. In adult melanoma, recurrent copy number gains at chromosome locations 1q, 6p, 7, 8q, 17q, 20q and deletions at 6q, 8p, 9p, 10q, have been observed [2, 9, 10-17]. In addition, some melanocytic neoplasms have their own signature gene abnormalities. For instance, Spitz nevi often present with HRAS mutations coupled with a gain in chromosome 11p whereas Spitzoid melanomas frequently have a homozygous deletion of 9p21 [10, 16, 18]. To date comparative genome hybridization, multiplex ligation probe amplification and fluorescent in situ hybridization (FISH) have been shown to have value in the differential diagnosis of Spitzoid tumor and melanoma in adults [2, 9, 14, 15, 19]. Few studies have investigated chromosomal abnormalities in pediatric melanomas. Several papers have reported 9p21 (CDKN2A) deletion is common in childhood melanoma and may predict aggressive behavior [2, 12, 20]. Gains in 6p25(RREB1), 11q13 (CCND1), 8q24 (MYC), 4q12 (KIT), and loss in 6q23 have also been observed but at different frequencies in Spitzoid melanoma relative to conventional melanoma [2]. Therefore, molecular tests could be potentially useful in diagnostically challenging cases of melanocytic lesions in children. NGS is the most recent molecular technique that has the potential to analyze the tumor genome and detect multiple gene abnormalities simultaneously. Recently, we had a case of childhood melanoma for which the molecular studies including NGS were employed to assist and confirm the diagnosis as reported here.
| Case Report | ▴Top |
A 12-year-old female with an unremarkable past medical history presented to her family doctor with a newly developed mole on the inner aspect of her right thigh. Her family history was significant for non-melanoma skin cancers in the grandparents. Physical examination revealed that she had fair-skin (Fitzpatrick type I) with multiple moles. The newly developed mole was slightly polypoid with an irregular border and uneven color, measuring 0.9 cm at the greatest dimension. A shave biopsy was performed.
Histopathological findings
Histology examination revealed a polypoid melanocytic proliferation in confluent nests distributing at all levels of the epidermis in a pagetoid fashion and in sheath-like growth expanding the dermis and extending to the base of the biopsy (Fig. 1). The tumor was composed of predominantly large epithelioid melanocytes showing sticking cytological atypia, lack of maturation and mitotic activity. The diagnosis of malignant melanoma was considered seriously, which would be a melanoma with at least a Clarke level of IV and 1.15 mm in Breslow thickness that requires wide re-excision and lymph node dissection. However, considering the patient’s young age, the differential diagnosis should include Spitz nevus which could have marked cytological atypia, pagetoid spread and mitotic activity. Nevertheless, the lack of maturation and the presence of deep dermal mitosis and parenchymal invasion suggested a malignant biological behavior for this melanocytic neoplasm.
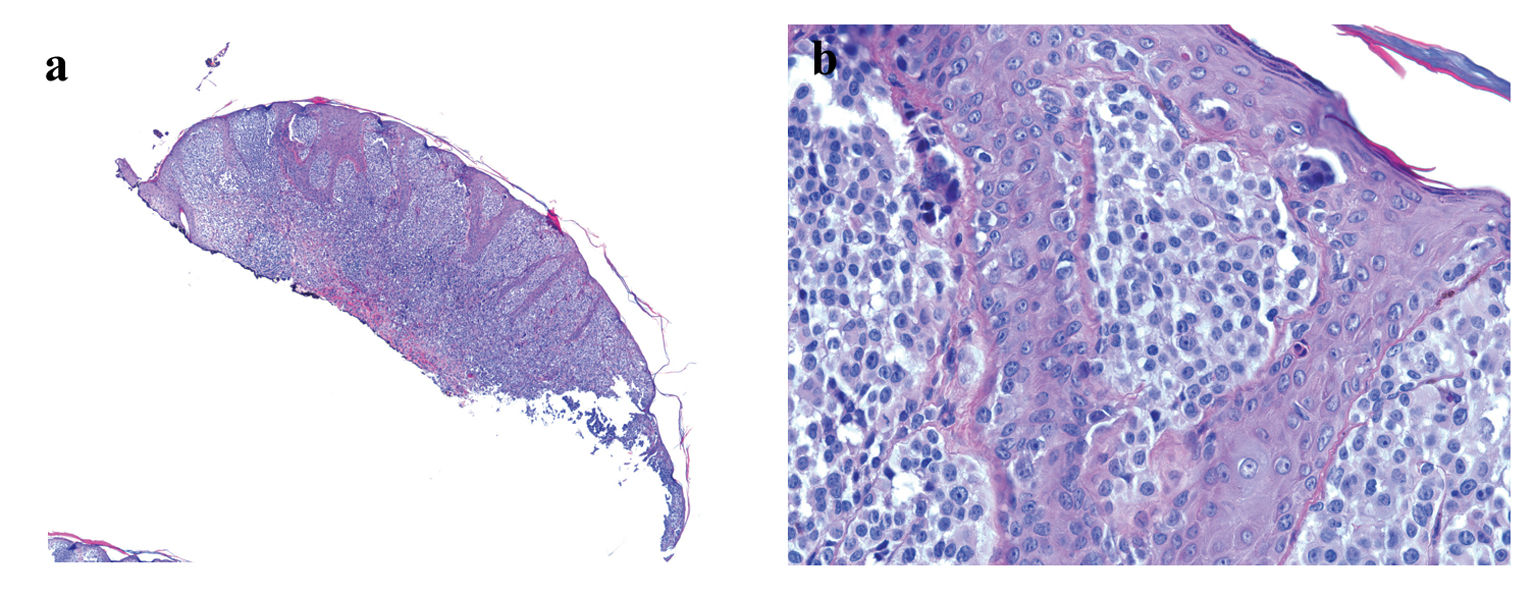 Click for large image | Figure 1. (a) Low-power image of H&E-stained section showing polypoid melanocytic neoplasm (H&E stain, × 40).(b) High-power image showing melanocytes with marked cytological atypia and pagetoid spread (H&E stain, × 400). |
Molecular studies
To provide further evidence regarding the real nature of this neoplasm, molecular studies of 12 genes frequently altered in melanoma were performed (Tables 1, 2). For NGS studies, the areas of the neoplastic and matched normal tissue were identified using H&E-stained slides macro-dissected from formalin-fixed, paraffin-embedded tissue. DNA was extracted using formalin fixed paraffin embedded tissue DNA extraction kit (Qiagen, Valencia, CA) and quantified by spectrometry (NanoDrop; Thermo Scientific, Waltham, MA) and fluorometry (Qubit; Life Technologies, Carlsbad, CA). An AmpliSeq Cancer Hotspot Panel v2 (Life Technologies) library was prepared for each sample. Amplicon libraries with barcodes were prepared from 10 ng template DNA using the Ion AmpliSeq Library Kit 2.0 (Life Technologies) and quantified by real-time PCR. Library pools were diluted, then combined together, mixed with IonSpheres (ISPs; Life Technologies) and subjected to emulsion PCR using the OT200 template kit (Life Technologies). Enrichment of ISPs with clonally amplified DNA was achieved using the OneTouch ES System (Life Technologies). Enriched ISPs were loaded onto an Ion 318 chip and sequenced using the ion sequencing kit (200 bp, Life Technologies).
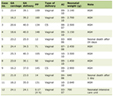 Click to view | Table 1. Top Gene Mutations in Melanocytic Lesions and NGS 12 Gene Panel for the Pediatric Melanoma Case |
Analysis of the raw data files consisted of three pipelines which included Variant Caller (Life Technologies), Ion Reporter (Life Technologies) and NextGen (Softgenetics). Results from the three software packages were compared and mutations detected were confirmed by Sanger sequencing. Mutation analysis revealed that the tumor was positive for a BRAF mutation (p.V600E), and negative for mutations in other genes (Table 1).
Copy number variation between the melanocytic neoplasm and matched normal tissue was examined using these software packages and in-house bioinformatics. For the in-house analysis, tumor amplicon read number (coverage) normalized to total number of tumor library reads was divided by matched normal read number normalized to total normal library reads. Amplicons with a read number below the 25% quartile for library reads were excluded from analysis. The tumor/normal ratio was divided by the average tumor/normal ratio for all 205 amplicons. A tumor/normal gene ratio greater than 1.2 was defined as a gain and a ratio less than 0.8 was defined as a loss. These cutoffs are based on 50% tumor cells with 1 or 3 copies of a gene in a background of normal cells. Copy number gains were observed in the BRAF (7q34) gene, consistent with 4 copies per cell and copy number deletions were observed in PTEN (10q23), CDKN2A (9p21), NRAS (1p13) consistent with 1 copy per cell.
To confirm gene copy number changes detected by NGS and extend the analysis to additional genes commonly used to distinguish benign nevi from malignant melanoma or Spitzoid melanoma, FISH was performed using an Abbott probe (Vysis, Des Plaines, IL). Probe sets included CCND1 (11q13), RREB1 (6p25), Cep 6 and MYB (6q23), previously reported as panel 1, CDKN2A (9p21), MYC (8q24) components of panel 2 and PIK3CA (3q26), PTEN (10q23), c-KIT (4q12) which have been implicated as recurrent abnormalities in other reports [2, 9-17, 21]. FISH analysis confirmed the BRAF (7q34) gain and the heterozygous deletion of PTEN (10q23), and CDKN2A (9p21). This melanocytic neoplasm also showed gains in the MYC (8q24) gene (Fig. 2).
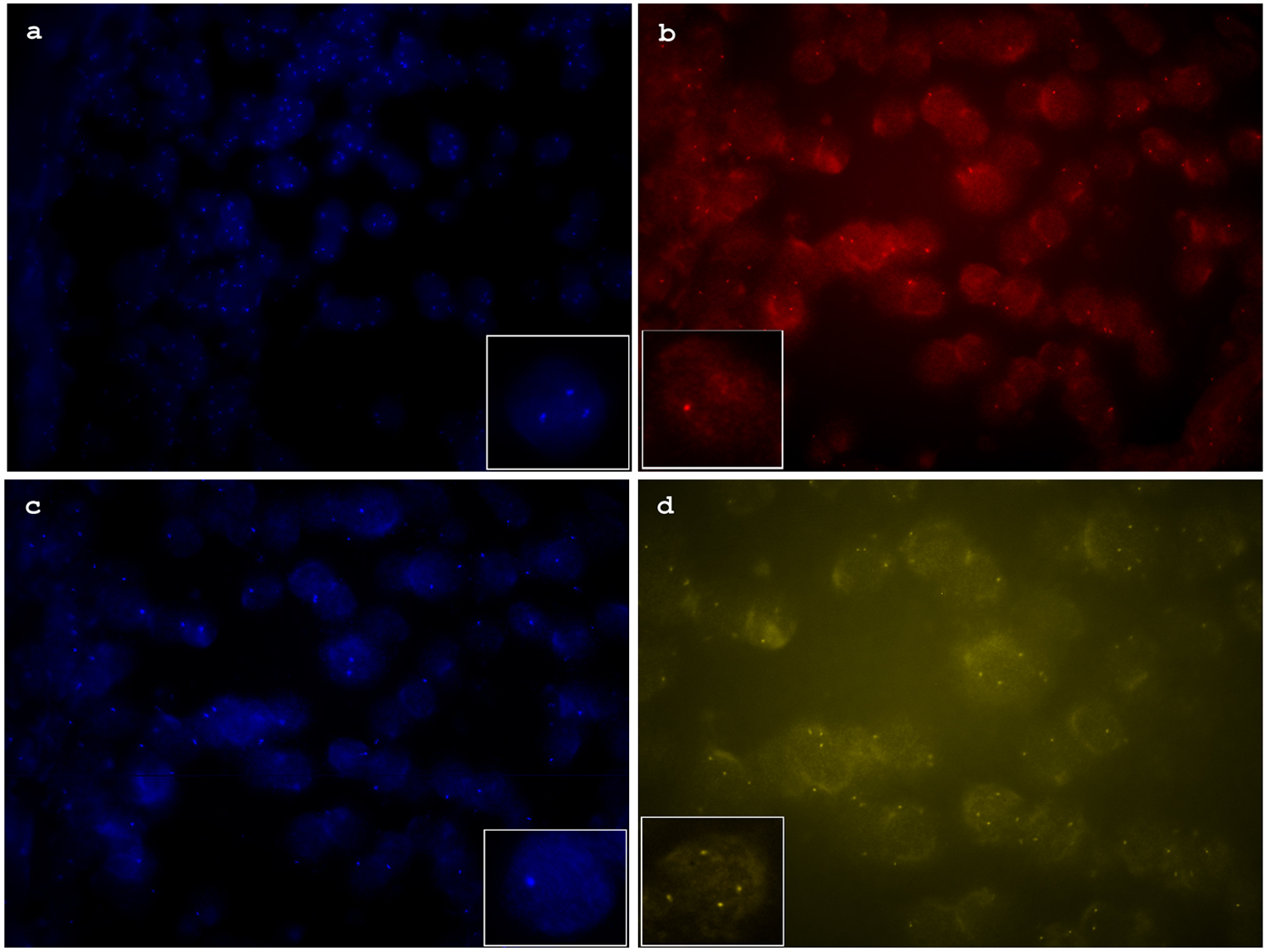 Click for large image | Figure 2. (a) Florescence in situ hybridization (FISH) image of aqua channel showing increased copy numbers of MYC (8q24). (b) FISH image of red channel showing heterozygous deletion of CDKN2A (9p21). (c) FISH image of aqua channel showing heterozygous deletion of PTEN (10p23). (d) FISH image of yellow channel showing increased copy numbers of BRAF (7p34). |
Based on histopathologic changes and the results of above molecular studies, a diagnosis of malignant melanoma was made. Following the pathologist’s advice, a wide local excision and sentinel lymph node biopsy were performed which showed residual melanoma with no lymph node metastasis. The patient recovered well from the surgery and was free of disease at 14-month follow-up.
| Discussion | ▴Top |
Malignant melanoma is deadly but fortunately rare in children. On the other hand, Spitz nevus is a benign melanocytic lesion mostly seen in children [8]. However, Spitz nevus and melanoma share many clinical as well as histopathologic similarities. Prior to 1950s, all the Spitz nevi were diagnosed as malignant melanomas before Dr. Spitz noted in her notorious study that most so called “childhood melanoma” had benign behavior and disease process. Her observation has been confirmed by many other studies and this kind of pediatric melanocytic tumor was thus relabeled as “Spitz nevus” to honor Dr. Spitz’s contribution [22]. Unfortunately, there has been a lack of objective criteria, both clinically and histopathologically, for the distinction of these two types of melanocytic tumors. This limitation has resulted in a high frequency of diagnostic discrepancy and misdiagnosis, with subsequent mismanagement of the patients [23].
As molecular analysis is increasingly being employed to aid in the diagnosis and choice of targeted therapy for cutaneous malignancies, ongoing efforts have being made to establish molecular markers that distinguish Spitz nevus from malignant melanoma. NGS has distinguished itself from other molecular studies by its ability to rapidly sequence large span of DNA across entire genomes. It can generate a few terabase (Tb) of data in a single sequencing run, which usually takes only 2 - 3 days [24]. In our case, we detected, in addition to a BRAF mutation, five chromosome abnormalities (BRAF (7q34), PTEN (10q23), CDKN2A (9p21), NRAS (1p13) and MYC (8q24)). The BRAF mutation was the first gene abnormality identified and targeted for therapy in melanoma. However, it is the most frequent mutation detected in both melanoma and benign nevi (50%), although less commonly in Spitz nevi (6%) and cannot be used as the sole criteria for melanoma diagnosis. HRAS (23%) is the most common mutation in Spitz nevi followed by BRAF and NRAS (Table 1). The copy number increase (7q34, 8q24) and decrease (9p21, 10q23) observed in this neoplasm are commonly observed in conventional melanoma. Recent evidence indicates CDKN2A (9p21) homozygous deletion is associated with aggressive clinical behavior in both pediatric and adult forms of Spitzoid melanoma [2, 14, 25]. By contrast, few copy number changes are seen in Spitz nevi other than gains in 11p and rarely in 7q [2, 9, 23]. Although no chromosome aberrations have been linked to tumor progression or malignant behavior in traditional pediatric melanoma, melanomas with malignant potential show multiple mutations and gene abnormalities such as deletions or duplications, while benign nevi have very few of these abnormalities [2, 6, 14, 15, 21]. Thus, the multiple gene abnormalities revealed in our case support the diagnosis of melanoma rather than Spitz nevus.
An FISH panel (CCND1, RREB1, Cep 6, MYB) has been used by many laboratories to distinguish histologically unequivocal melanomas from benign nevi with a reported sensitivity of 75-86.7% for conventional melanoma and 70% for Spitzoid melanoma [9]. No copy number aberrations were detected with this the traditional FISH panel in our case. We also used an updated FISH panel consisting of PIK3CA, RREB1, CDKN2A and MYC with a reported sensitivity of 94% [14]. In our case, this updated FISH panel did reveal a homozygous CDKN2A (9p21) deletion which is one of the most common chromosome abnormalities in adult or pediatric Spitzoid melanomas [2]. But given our findings, which include the MYC gain, the chromosome pattern in this melanocytic neoplasm is most consistent with a conventional melanoma rather than Spitzoid melanoma.
In summary, we present the usage of NGS and other molecular studies in assisting diagnostically challenging case of a pediatric melanoma. Although histopathologic examination favored the diagnosis of malignant melanoma, molecular studies helped to confirm the diagnosis. Furthermore, we showed that NGS can detect both gene mutation and the gene number variation in a large gene panel from a limited specimen. However, this method requires an abundant tumor component (50%) or micro-dissection is required. When combined with recent melanoma FISH panels, which allows direct visualization of genes at the individual cell level, this approach can improve the confidence of a diagnosis. We believe that the integration of molecular diagnostic information with histopathologic features contributes significantly toward accurate diagnosis and potential targeted therapy for pediatric melanocytic lesions.
Acknowledgments
We want to thank Karen Dresser for her assistance in archived tissue block retrieval and section.
Disclosures
None declared.
| References | ▴Top |
- Bis S, Tsao H. Melanoma genetics: the other side. Clin Dermatol. 2013;31(2):148-155.
doi pubmed - Gerami P, Cooper C, Bajaj S, Wagner A, Fullen D, Busam K, Scolyer RA, et al. Outcomes of atypical spitz tumors with chromosomal copy number aberrations and conventional melanomas in children. Am J Surg Pathol. 2013;37(9):1387-1394.
doi pubmed - Mills O, Messina JL. Pediatric melanoma: a review. Cancer Control. 2009;16(3):225-233.
- Miteva M, Lazova R. Spitz nevus and atypical spitzoid neoplasm. Semin Cutan Med Surg. 2010;29(3):165-173.
doi pubmed - Wechsler J, Bastuji-Garin S, Spatz A, Bailly C, Cribier B, Andrac-Meyer L, Vergier B, et al. Reliability of the histopathologic diagnosis of malignant melanoma in childhood. Arch Dermatol. 2002;138(5):625-628.
doi pubmed - Barnhill RL, Flotte TJ, Fleischli M, Perez-Atayde A. Cutaneous melanoma and atypical Spitz tumors in childhood. Cancer. 1995;76(10):1833-1845.
doi - Kapur P, Selim MA, Roy LC, Yegappan M, Weinberg AG, Hoang MP. Spitz nevi and atypical Spitz nevi/tumors: a histologic and immunohistochemical analysis. Mod Pathol. 2005;18(2):197-204.
doi pubmed - Chao MM, Schwartz JL, Wechsler DS, Thornburg CD, Griffith KA, Williams JA. High-risk surgically resected pediatric melanoma and adjuvant interferon therapy. Pediatr Blood Cancer. 2005;44(5):441-448.
doi pubmed - Gerami P, Jewell SS, Morrison LE, Blondin B, Schulz J, Ruffalo T, Matushek Pt, et al. Fluorescence in situ hybridization (FISH) as an ancillary diagnostic tool in the diagnosis of melanoma. Am J Surg Pathol. 2009;33(8):1146-1156.
doi pubmed - Bastian BC, Olshen AB, LeBoit PE, Pinkel D. Classifying melanocytic tumors based on DNA copy number changes. Am J Pathol. 2003;163(5):1765-1770.
doi - Bastian BC, Wesselmann U, Pinkel D, Leboit PE. Molecular cytogenetic analysis of Spitz nevi shows clear differences to melanoma. J Invest Dermatol. 1999;113(6):1065-1069.
doi pubmed - Daniotti M, Ferrari A, Frigerio S, Casieri P, Miselli F, Zucca E, Collini P, et al. Cutaneous melanoma in childhood and adolescence shows frequent loss of INK4A and gain of KIT. J Invest Dermatol. 2009;129(7):1759-1768.
doi pubmed - Dutton-Regester K, Hayward NK. Reviewing the somatic genetics of melanoma: from current to future analytical approaches. Pigment Cell Melanoma Res. 2012;25(2):144-154.
doi pubmed - Gerami P, Li G, Pouryazdanparast P, Blondin B, Beilfuss B, Slenk C, Du J, et al. A highly specific and discriminatory FISH assay for distinguishing between benign and malignant melanocytic neoplasms. Am J Surg Pathol. 2012;36(6):808-817.
doi pubmed - Gerami P, Scolyer RA, Xu X, Elder DE, Abraham RM, Fullen D, Prieto VG, et al. Risk assessment for atypical spitzoid melanocytic neoplasms using FISH to identify chromosomal copy number aberrations. Am J Surg Pathol. 2013;37(5):676-684.
doi pubmed - Shahbain H, Cooper C, Gerami P. Molecular diagnostics for ambiguous melanocytic tumors. Semin Cutan Med Surg. 2012;31(4):274-278.
doi pubmed - Woodman SE, Lazar AJ, Aldape KD, Davies MA. New strategies in melanoma: molecular testing in advanced disease. Clin Cancer Res. 2012;18(5):1195-1200.
doi pubmed - McCalmont TH, Vemula S, Sands P, Bastian BC. Molecular-microscopical correlation in dermatopathology. J Cutan Pathol. 2011;38(4):324-326, 323.
- van Dijk MC, Rombout PD, Boots-Sprenger SH, Straatman H, Bernsen MR, Ruiter DJ, Jeuken JW. Multiplex ligation-dependent probe amplification for the detection of chromosomal gains and losses in formalin-fixed tissue. Diagn Mol Pathol. 2005;14(1):9-16.
doi pubmed - Vignoli M, Scaini MC, Ghiorzo P, Sestini R, Bruno W, Menin C, Gensini F, et al. Genomic rearrangements of the CDKN2A locus are infrequent in Italian malignant melanoma families without evidence of CDKN2A/CDK4 point mutations. Melanoma Res. 2008;18(6):431-437.
doi pubmed - Kraehn GM, Utikal J, Udart M, Greulich KM, Bezold G, Kaskel P, Leiter U, et al. Extra c-myc oncogene copies in high risk cutaneous malignant melanoma and melanoma metastases. Br J Cancer. 2001;84(1):72-79.
doi pubmed - Spitz S. Melanomas of childhood. Am J Pathol. 1948;24(3):591-609.
pubmed - Pimiento JM, Larkin EM, Smalley KS, Wiersma GL, Monks NR, Fedorenko IV, Peterson CA, et al. Melanoma genotypes and phenotypes get personal. Lab Invest. 2013;93(8):858-867.
doi pubmed - An Introduction to Next-Generation Technology. N.p., n.d. Web. 25 Dec 2013. <http://res.illumina.com/documents/products>.
- Stark M, Hayward N. Genome-wide loss of heterozygosity and copy number analysis in melanoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2007;67(6):2632-2642.
This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
International Journal of Clinical Pediatrics is published by Elmer Press Inc.
